
Research
Research focuses on the identification of clinically relevant molecular subgroups and functional characterization of genes involved in glioma initiation and/or progression using state of the art high-throughput genomic approaches. Identification of clinically relevant molecular subgroups Correct classification of gliomas is important because it directly affects the path of treatment to be followed. However, classification of gliomas based on histological appearance alone can be difficult and subject to significant inter-observer variability. The WHO 2021 classification of tumors of the central nervous system therefore incorporates specific molecular markers for tumour diagnosis. However, even within these molecularly defined entities, some patients have a better prognosis than others. Identification of molecular markers of tumor malignancy therefore is a key aim of our lab. Identification of such malignancy markers is done by:
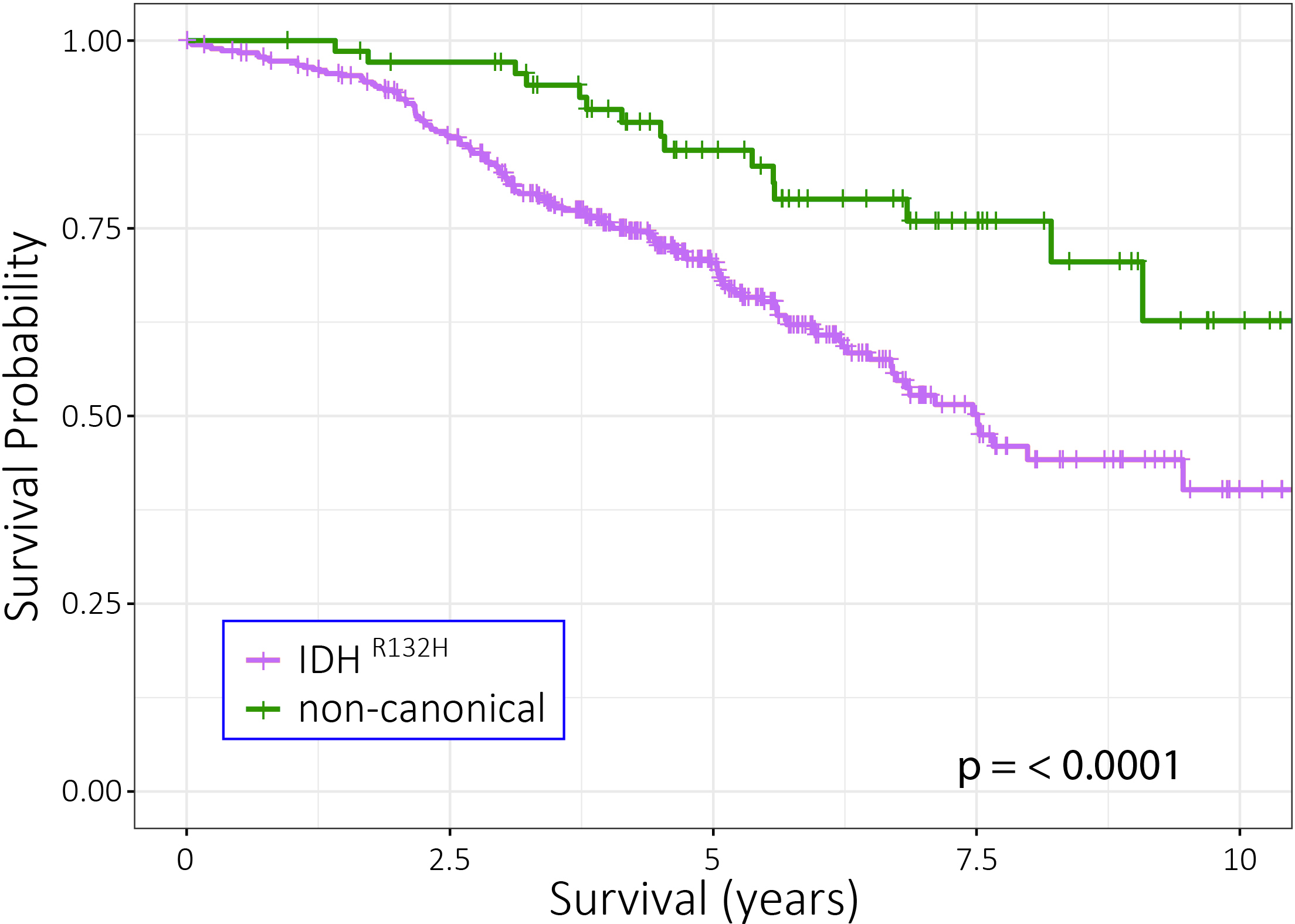 Figure 1: Astrocytoma patients with non-canonical IDH mutations have better outcome than patients harbouring tumors with the canonical IDH1-R132H mutation. Functional analysis of genes involved in glioma initiation and/or progression Genetic changes involved in glioma-genesis or progression are potential therapeutic targets and a number of such changes have been identified in gliomas. In GBMs, causal genes include the oncogenes EGFR, MDM2, and CDK4 and the tumor suppressor genes PTEN, CDKN2A and NF1. In lower grade gliomas, causal cancer genes include IDH1, TP53, CIC, FUBP1, ATRX and NOTCH1. We are currently employing next generation sequencing (bulk DNA and RNA-seq, as well as single cell/nucleus sequencing and spatial transcriptomics) in combination with other state of the art 'omics' techniques (including high-content, high-throughput confocal imaging, mass spectrometry and reverse phase protein arrays) to functionally characterize the causal genes of the various subtypes of gliomas. At present, the functional analysis is focussed on mutations in the IDH1 and EGFR genes. Our aim is to determine the role of these proteins in glioma initiation and/or progression, explain why specific mutations accumulate in distinct tumor subtypes and identify novel drugs that target these genetic changes. For example, the epidermal growth factor receptor (EGFR) gene is a key oncogene that is mutated in many tumors including lung adenocarcinomas (LUAD) and glioblastomas (GBM). In LUAD, around 90% of all EGFR mutations comprise of either short in-frame deletions in exon 19 (in particular around residues 747-750) or the L858R missense mutation in exon 21. In GBMs, the EGFR gene is also mutated, but here the gene is high copy number amplified with levels sometimes exceeding 100 copies per cell. This initial amplification is then followed by the acquisition of additional mutations that including intragenic deletions, point mutations and gene-fusions. 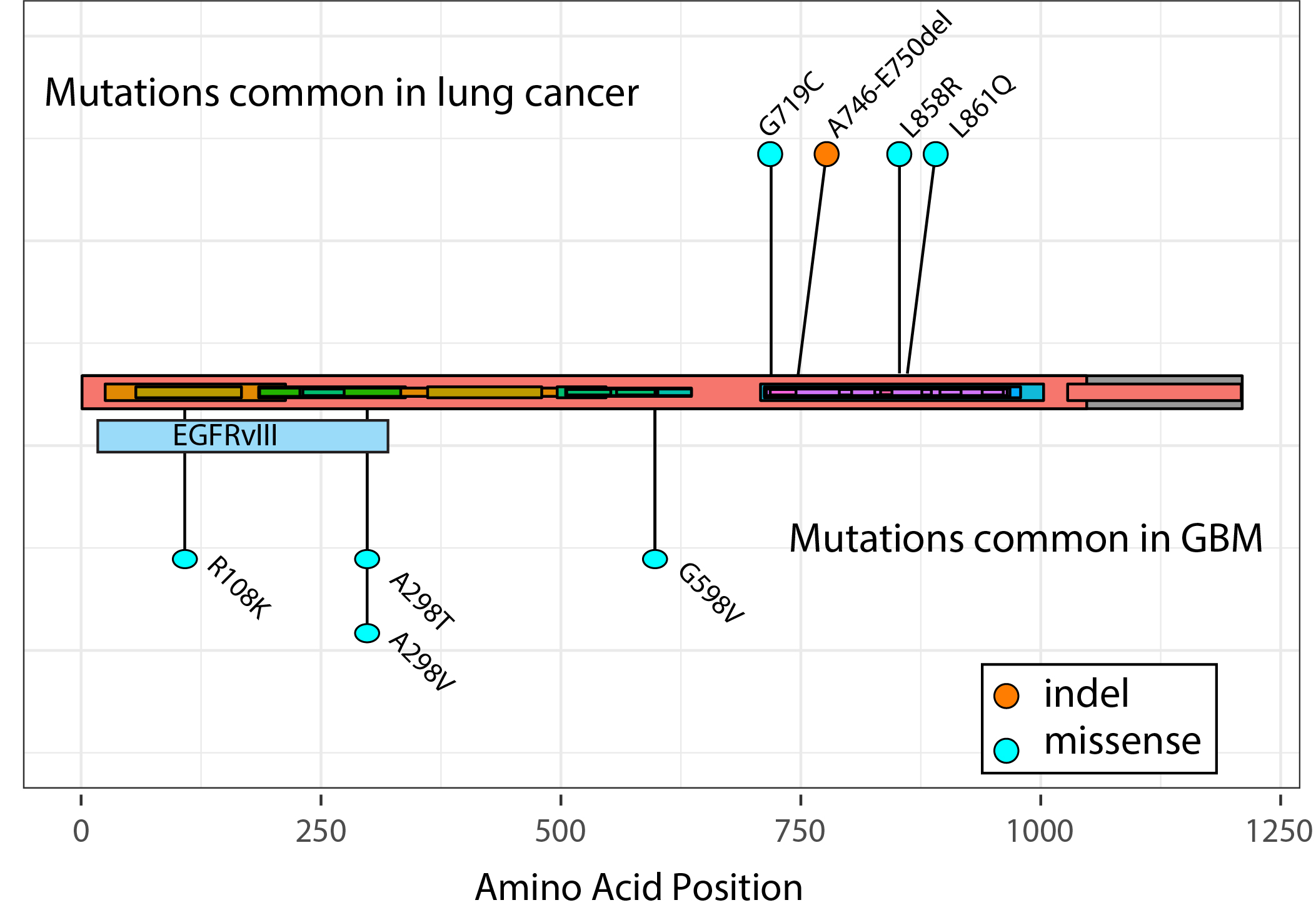 Figure 2: Distribution of EGFR mutations in glioblastomas and lung cancer. As can be seen, each tumor-type has its own distribution of mutations in EGFR with those in GBMs concentrating on the C-terminal extracellular domain. EGFRvIII is the most common mutation in GBMs, present in ~50% of tumors, and is an in-frame deletion of exons 2-7.
|
 |